Using the new method of 19F chemical shift fingerprinting to identify the topological conformations of long telomere G4
G4 is a non-canonical nucleic acid secondary structure formed by G-rich DNA or RNA, which is ubiquitous in living organisms and has important biological functions. Human telomere DNA is composed of a tandem repeat of 5-15 kb of the hexanucleotide sequence TTAGGG, with a single-stranded overhang at the 3' end ranging from 50-300 nucleotides. Under physiological conditions, it can fold into multiple G4 structures. These special structures play an important role in maintaining telomere ends and are also considered potential targets for anticancer drugs. Therefore, clarifying the folding details of telomere 3' overhang G4, including topological conformational information and relative proportions, is crucial for understanding telomere function and the development of anticancer drugs targeting telomere G4. However, due to the large molecular weight of naturally long telomere overhangs, the coexistence of multiple G4 structures in a conformational dynamic equilibrium, and the complex intracellular environment, traditional structural characterization methods such as conventional NMR, X-ray crystallography, and circular dichroism spectroscopy are not applicable. This has led to the inability to confirm the structural information of naturally long human telomere overhangs in dilute solutions and physiological conditions.
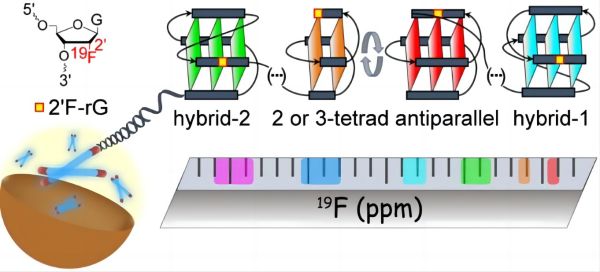
19F NMR has unique advantages in characterizing human telomere G4 structures due to its natural abundance, high signal sensitivity, lack of intracellular background interference, and strong conformational dependence of chemical shifts. By introducing 19F labeling at specific sites in the nucleic acid sequence, the research team found that different topological structures have characteristic chemical shift distribution ranges. Based on this, they summarized the 19F chemical shift fingerprinting for human telomere G4 topologies. Using this fingerprinting technique, the research team systematically studied the G4 conformational information of naturally long human telomere sequences and found that long telomeres tend to form stable G4 at the 5' and 3' ends, while the internal G4 are relatively more dynamic. The redundant repeats that do not form G4 exist in an unfolded form within the sequence. All G4 are in a conformational dynamic equilibrium, with the main topological conformations of the 5' and 3' terminal G4 being hybrid-2 and hybrid-1, respectively. The internal G4 are mainly 2 or 3-layer antiparallel conformations, and these G4 conformations are independent of each other. The research team further studied the structure of long telomere sequences in African clawed frog oocytes and found that the conformational details were consistent with those in K+ dilute solution, and were not disturbed by complex cellular environments such as crowding. They also found that the stability of telomeres in live cells is regulated by sequence length, suggesting that telomere length may affect protein accessibility and thereby regulate telomere function. This work provides an effective method for characterizing the topological structure of naturally long human telomere overhangs, and presents the most detailed topological structural model to date. It provides an important structural basis for understanding telomere structure, function, and the development of anticancer drugs targeting telomere G4. At the same time, the concept of fingerprinting is also expected to be extended to other nucleic acid secondary structural systems.
The research was published in the academic journal "Journal of the American Chemical Society"(JACS) under the title “19F Nuclear Magnetic Resonance Fingerprinting Technique for Identifying and Quantifying G-Quadruplex Topology in Human Telomeric Overhangs.” APM was the first completion unit for this work. WANG Chen, a postdoctoral researcher from APM, was the first author, and LI Conggang, a researcher, was the corresponding author.
This research work was funded by the National Natural Science Foundation of China, the Ministry of Science and Technology, and the Chinese Academy of Sciences.
Link to the article:
