Recently, a collaborative team from the Complex Biological Systems Spectroscopy Analysis Group at the Innovation Academy for Precision Measurement Science and Technology (APM) and the Mass Spectrometry Analysis Group at the Dalian Institute of Chemical Physics achieved significant methodological breakthroughs in in-cell protein structure research. By systematically evaluating the performance of multiple in situ chemical cross-linkers in native cellular environments, the researchers elucidated the relationship between cross-linker structural rigidity and their capacity to capture protein dynamic information, thereby establishing a theoretical foundation for efficient investigation of protein dynamics and interactions. These findings were recently published in JACS Au.
Protein structure and its conformational dynamics constitute the cornerstone for understanding biological function. Current experimental studies are predominantly confined to analyzing purified proteins in vitro, whereas structure prediction methods like AlphaFold are limited to predicting static protein conformations, failing to capture the dynamic structural rearrangements occurring within cellular environments. In situ chemical cross-linking mass spectrometry (CX-MS), as an emerging technology, enables the investigation of protein structures, dynamics, and interactions within cellular contexts. However, a systematic understanding of how the structural properties of distinct cross-linkers influence their capacity to capture protein dynamic information in cellular environments has long been lacking.
This study conducted a comprehensive comparative evaluation of the intracellular performance of four mainstream in situ chemical cross-linkers. By integrating protein structural analysis with all-atom molecular dynamics simulations, we systematically elucidated the relationships between structural parameters (including cross-linker length, backbone rigidity, and dihedral angles) and the dynamic properties of cross-linkers in cellular environments. The results revealed that while cross-linkers of varying lengths exhibited comparable efficiency in capturing intra-protein amino acid distances, this observed 'convergence' did not originate from complete extension of the cross-linkers, but was instead strongly correlated with their intrinsic dynamic properties. This finding fundamentally redefines the conventional paradigm of cross-linker performance by demonstrating that dynamic behavior, rather than static structural features, governs cross-linking efficacy in cellular environments.
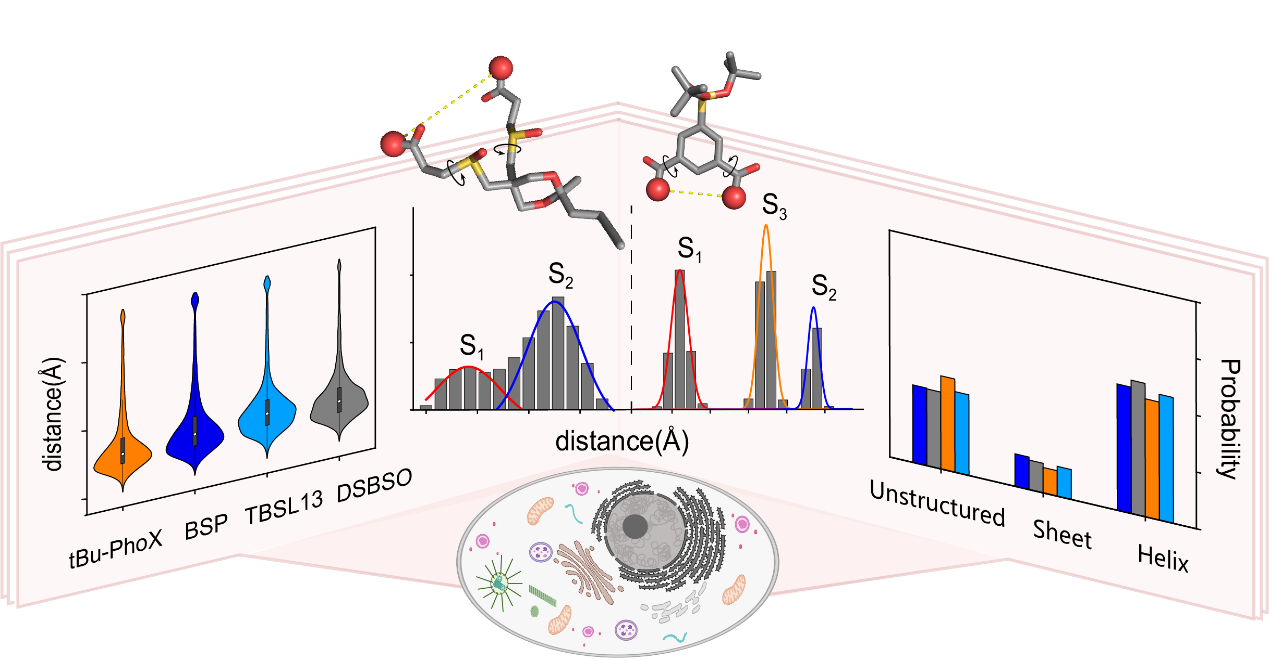
The dynamic properties of different cross-linkers determine their capacity for probing protein structures in cellular environments
Crucially, the research team demonstrated that cross-linkers with enhanced backbone rigidity exhibit significantly higher efficiency in capturing structural information from intrinsically disordered regions (IDRs) of proteins and their dynamic conformational ensembles. If chemical cross-linking is analogized as a molecular ruler for probing protein architecture, rigid cross-linkers resemble precision 'straight rulers' with defined geometric constraints, whereas flexible counterparts function as bendable 'tape measures'—though the latter exhibit superior structural adaptability, the former confer distinct advantages in resolving dynamic conformational landscapes. This study not only provides robust data support for intracellular cross-linking mass spectrometry (XL-MS) experiments but will also propel advancements in the field of cellular structural biology.
The research findings were published in JACS Au under the title "Dissecting in vivo Cross-Linker Performance: Insights into Structural Dynamics and Molecular Rigidity". Ph.D. students ZHOU Yuxuan at APM and ZHANG Beirong at the Dalian Institute of Chemical Physics served as co-first authors. Associate professor GONG Zhou at APM and professor ZHAO Qun at the Dalian Institute of Chemical Physics are the corresponding authors.
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, the National Key Research and Development Program of China, and the National Natural Science Foundation of China (NSFC).
Link to the article: https://pubs.acs.org/doi/10.1021/jacsau.5c00709
